5.5 EMP_impute
Data imputation is estimating or populating missing values in a dataset by an algorithm when those missing values exist. The goal is to provide reasonable estimates of where missing values are located so that these data can be better utilized in subsequent data analysis or modeling. The module EMP_impute performs data interpolation based on the Chained Random Forests (CRF) algorithm. This algorithm is more scientific than the traditional mean or mode interpolation method. It can be used not only to impute continuous variables but also to impute categorical variables. ModuleEMP_impute supports data interpolation for assay, rowdata, and coldata.
5.5.1 Impute coldata (Sample-Related Data)
Coldata of the project refers to sample-related data, which often have different degrees of missing values. Common causes of missing values include errors or omissions in the data collection process, withdrawal of subjects from the study, technical problems (e.g., equipment failure or data transmission errors), etc. The module EMP_impute can impute missing values of coldata based on the CRF algorithm.
🏷️Example:
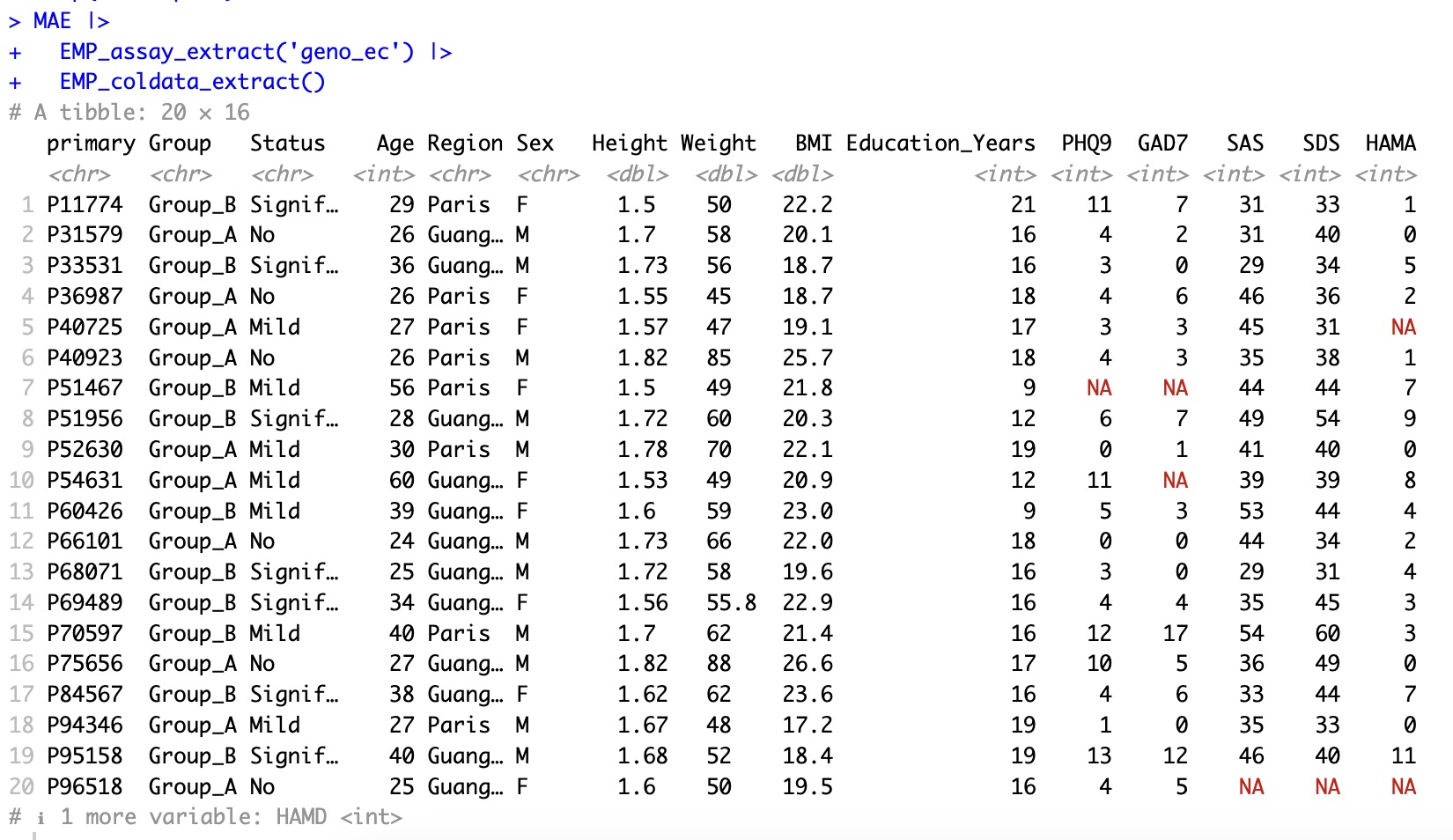
Before imputation: coldata has lots of missing values.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_coldata_extract()
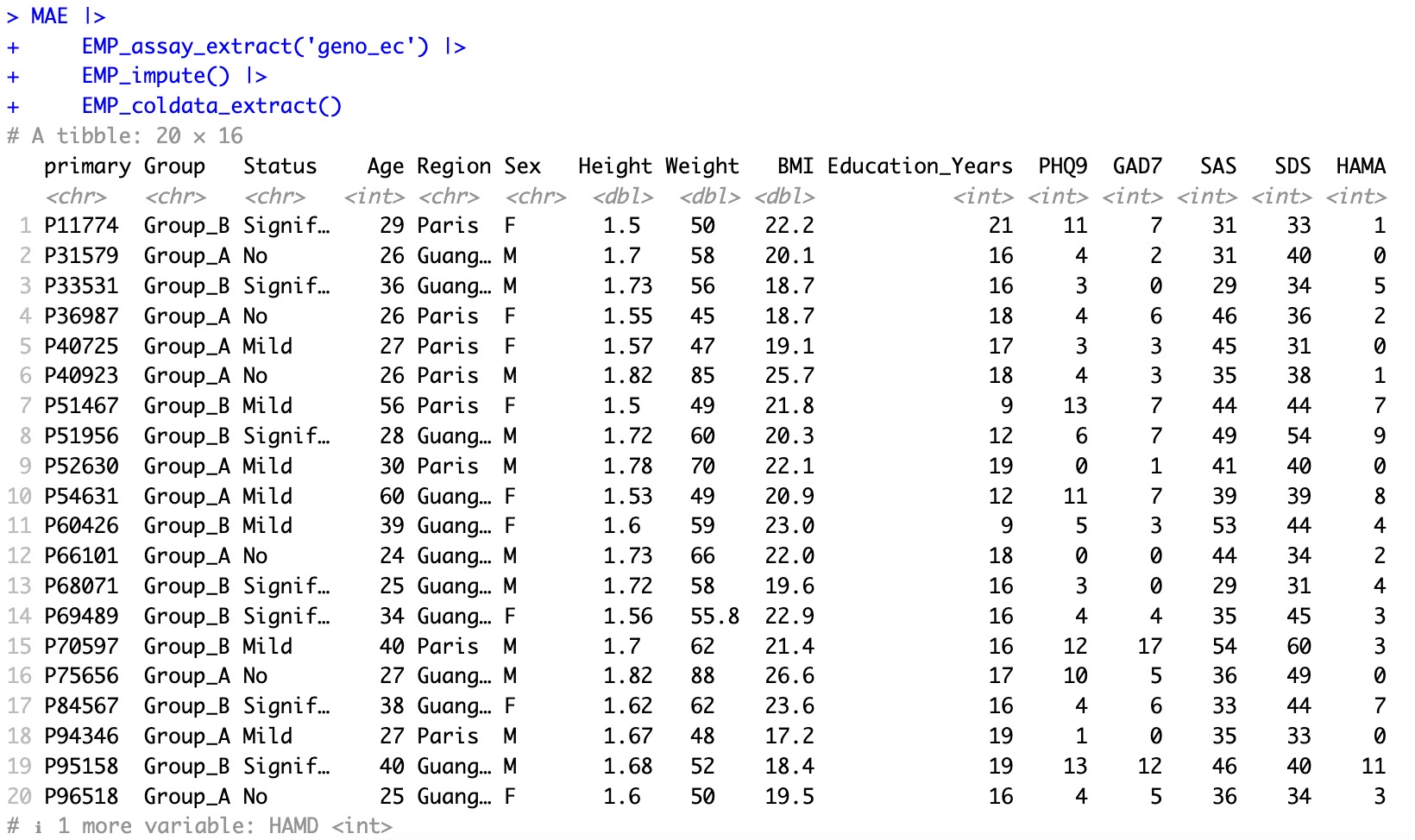
After imputation: impute all missing values of coldata.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_impute() |>
EMP_coldata_extract()
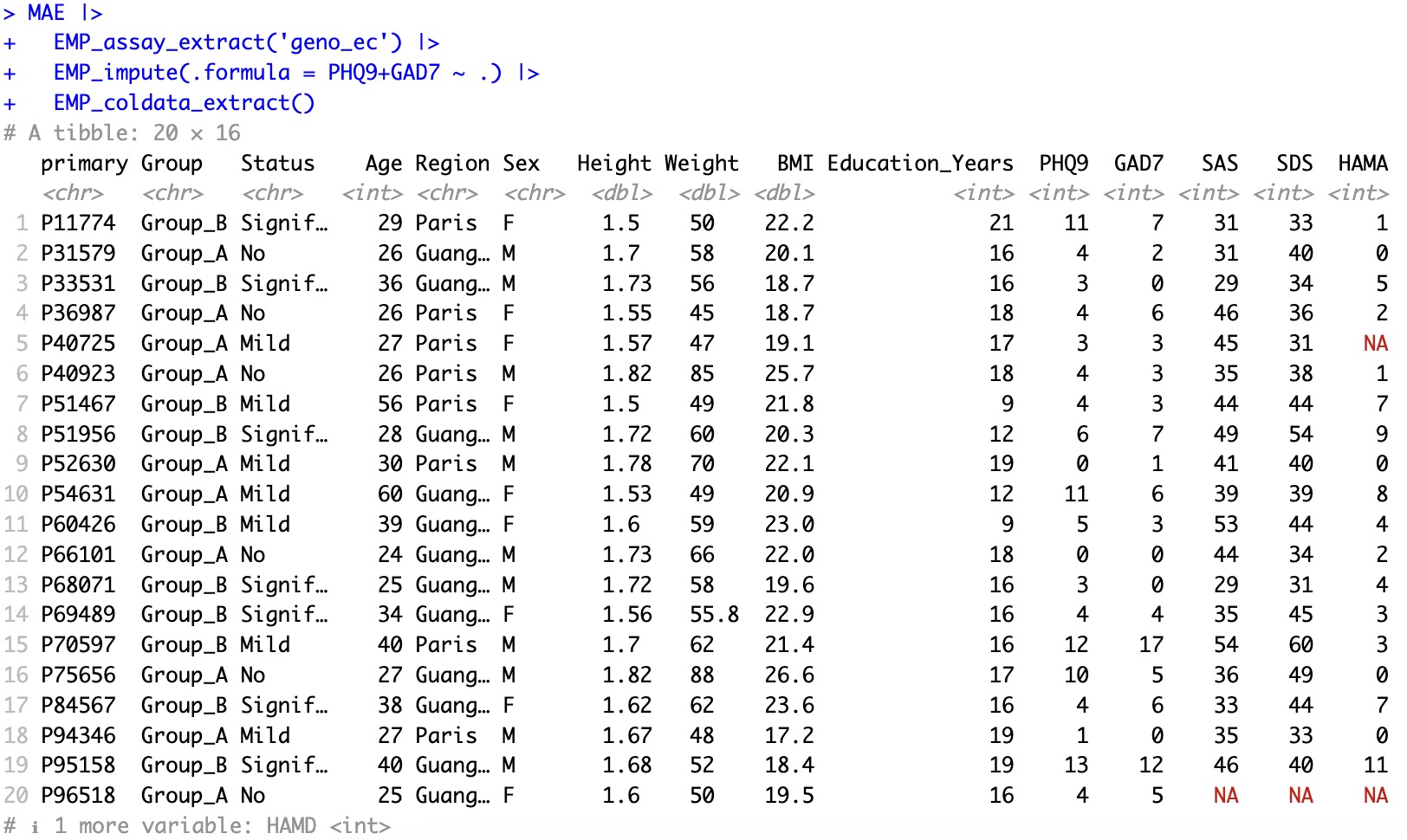
Users can also impute only partial missing values of coldata. For example, only missing values for PHQ9 and GAD7 are imputed.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_impute(.formula = PHQ9+GAD7 ~ .) |>
EMP_coldata_extract()
5.5.2 Impute assay (Experimental data)
When there are no missing values in the assay, imputation using the module EMP_impute prompts "Assay data has no NA value! ".

5.5.3 Impute rowdata (feature-related data)
Rowdata of the project refers to feature-related data, and its missing values are mainly caused by imperfect database annotations. Therefore, although the module EMP_impute supports the interpolation of rowdata using the parameterrowdata=T, the interpolation result is difficult to meet the actual needs, so it is not recommended to impute rowdata.
5.5.4 Imputation of the entire multi-omics object's coldata
Users can also directly perform phenotype data imputation on the entire multi-omics data container. It should be noted that in this case, the output object will be a MultiAssayExperiment.
MAE |>
EMP_impute()
